Activations and Initializations#
In this course, we will revisit the Fully Connected model presented in Course 5 on NLP. We will study the behavior of activations throughout the network during its initialization. This course is inspired by Andrej Karpathy’s course, titled Building makemore Part 3: Activations & Gradients, BatchNorm.
Neural networks have several advantages:
They are very flexible and can solve many problems.
They are relatively simple to implement.
However, they can be complex to optimize, especially when dealing with deep networks.
Code Review#
We are revisiting the code from Notebook 3 of Course 5 on NLP.
import torch
import torch.nn.functional as F
import matplotlib.pyplot as plt
%matplotlib inline
words = open('../05_NLP/prenoms.txt', 'r').read().splitlines()
chars = sorted(list(set(''.join(words))))
stoi = {s:i+1 for i,s in enumerate(chars)}
stoi['.'] = 0
itos = {i:s for s,i in stoi.items()}
For educational purposes, we will not use PyTorch’s dataset and dataloader. We will evaluate the loss at the beginning of training after the first batch. Overall, it works the same way, except that we take a random batch at each iteration instead of going through the entire dataset at each epoch.
block_size = 3 # Contexte
def build_dataset(words):
X, Y = [], []
for w in words:
context = [0] * block_size
for ch in w + '.':
ix = stoi[ch]
X.append(context)
Y.append(ix)
context = context[1:] + [ix]
X = torch.tensor(X)
Y = torch.tensor(Y)
print(X.shape, Y.shape)
return X, Y
import random
random.seed(42)
random.shuffle(words)
n1 = int(0.8*len(words))
n2 = int(0.9*len(words))
Xtr, Ytr = build_dataset(words[:n1]) # 80%
Xdev, Ydev = build_dataset(words[n1:n2]) # 10%
Xte, Yte = build_dataset(words[n2:]) # 10%
torch.Size([180834, 3]) torch.Size([180834])
torch.Size([22852, 3]) torch.Size([22852])
torch.Size([22639, 3]) torch.Size([22639])
embed_dim=10 # Dimension de l'embedding de C
hidden_dim=200 # Dimension de la couche cachée
C = torch.randn((46, embed_dim))
W1 = torch.randn((block_size*embed_dim, hidden_dim))
b1 = torch.randn(hidden_dim)
W2 = torch.randn((hidden_dim, 46))
b2 = torch.randn(46)
parameters = [C, W1, b1, W2, b2]
for p in parameters:
p.requires_grad = True
max_steps = 200000
batch_size = 32
lossi = []
for i in range(max_steps):
# Permet de construire un mini-batch
ix = torch.randint(0, Xtr.shape[0], (batch_size,))
# Forward
Xb, Yb = Xtr[ix], Ytr[ix]
emb = C[Xb]
embcat = emb.view(emb.shape[0], -1)
hpreact = embcat @ W1 + b1
h = torch.tanh(hpreact)
logits = h @ W2 + b2
loss = F.cross_entropy(logits, Yb)
# Retropropagation
for p in parameters:
p.grad = None
loss.backward()
# Mise à jour des paramètres
lr = 0.1 if i < 100000 else 0.01 # On descend le learning rate d'un facteur 10 après 100000 itérations
for p in parameters:
p.data += -lr * p.grad
if i % 10000 == 0:
print(f'{i:7d}/{max_steps:7d}: {loss.item():.4f}')
lossi.append(loss.log10().item())
0/ 200000: 21.9772
10000/ 200000: 2.9991
20000/ 200000: 2.5258
30000/ 200000: 1.9657
40000/ 200000: 2.4326
50000/ 200000: 1.7670
60000/ 200000: 2.1324
70000/ 200000: 2.4160
80000/ 200000: 2.2237
90000/ 200000: 2.3905
100000/ 200000: 1.9304
110000/ 200000: 2.1710
120000/ 200000: 2.3444
130000/ 200000: 2.0970
140000/ 200000: 1.8623
150000/ 200000: 1.9792
160000/ 200000: 2.4602
170000/ 200000: 2.0968
180000/ 200000: 2.0466
190000/ 200000: 2.3746
plt.plot(lossi)
[<matplotlib.lines.Line2D at 0x7f028467b990>]
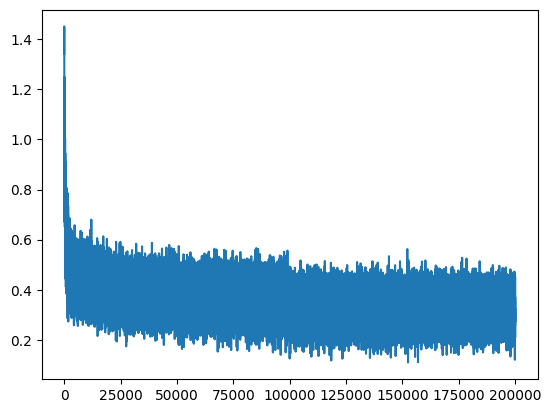
There is a lot of “noise” because we calculate the loss each time on small batches relative to the entire training dataset.
Abnormally High Loss at Initialization#
Training proceeds correctly. However, we notice something strange: the loss at the beginning of training is abnormally high. We would expect to get a value corresponding to a case where each letter has a uniform probability of occurrence (i.e., \(\frac{1}{46}\)).
In this case, the negative log likelihood would be: \(-ln(\frac{1}{46})=3.83\)
Therefore, it would be logical to obtain a value of this magnitude during the first loss calculation.
Small Example Illustrating the Problem#
To understand what is happening, let’s use a small example and observe the loss values depending on the initialization. Imagine that all the weights in logits are initialized to 0. In this case, we would obtain uniform probabilities.
logits=torch.tensor([0.0,0.0,0.0,0.0])
probs=torch.softmax(logits,dim=0)
loss= -probs[1].log()
probs,loss
(tensor([0.2500, 0.2500, 0.2500, 0.2500]), tensor(1.3863))
However, it is not recommended to initialize the weights of a neural network to 0. We used a random initialization based on a standard normal Gaussian distribution.
logits=torch.randn(4)
probs=torch.softmax(logits,dim=0)
loss= -probs[1].log()
probs,loss
(tensor([0.3143, 0.0607, 0.3071, 0.3178]), tensor(2.8012))
The problem becomes quickly apparent: the randomness of the Gaussian distribution tips the balance one way or the other (you can run the previous code several times to confirm this).
So, what can we do? Simply multiply our logit vector by a small value to decrease the initial weight values and make the softmax more uniform.
logits=torch.randn(4)*0.01
probs=torch.softmax(logits,dim=0)
loss= -probs[1].log()
probs,loss
(tensor([0.2489, 0.2523, 0.2495, 0.2493]), tensor(1.3772))
We obtain, more or less, the same loss as for uniform probabilities.
Note: However, we can initialize the bias value to zero, as it makes no sense to have a positive or negative bias at initialization.
Training with Initialization Adjustment#
Let’s revisit the previous code, but with the new initialization values.
C = torch.randn((46, embed_dim))
W1 = torch.randn((block_size*embed_dim, hidden_dim))*0.01 # On initialise les poids à une petite valeur
b1 = torch.randn(hidden_dim) *0 # On initialise les biais à 0
W2 = torch.randn((hidden_dim, 46))*0.01
b2 = torch.randn(46)*0
parameters = [C, W1, b1, W2, b2]
for p in parameters:
p.requires_grad = True
lossi = []
for i in range(max_steps):
ix = torch.randint(0, Xtr.shape[0], (batch_size,))
Xb, Yb = Xtr[ix], Ytr[ix]
emb = C[Xb]
embcat = emb.view(emb.shape[0], -1)
hpreact = embcat @ W1 + b1
h = torch.tanh(hpreact)
logits = h @ W2 + b2
loss = F.cross_entropy(logits, Yb)
for p in parameters:
p.grad = None
loss.backward()
lr = 0.1 if i < 100000 else 0.01
for p in parameters:
p.data += -lr * p.grad
if i % 10000 == 0:
print(f'{i:7d}/{max_steps:7d}: {loss.item():.4f}')
lossi.append(loss.log10().item())
0/ 200000: 3.8304
10000/ 200000: 2.4283
20000/ 200000: 2.0651
30000/ 200000: 2.1124
40000/ 200000: 2.3158
50000/ 200000: 2.2752
60000/ 200000: 2.1887
70000/ 200000: 2.1783
80000/ 200000: 1.8120
90000/ 200000: 2.3178
100000/ 200000: 2.0973
110000/ 200000: 1.8992
120000/ 200000: 1.6917
130000/ 200000: 2.2747
140000/ 200000: 1.8054
150000/ 200000: 2.3569
160000/ 200000: 2.4231
170000/ 200000: 2.0711
180000/ 200000: 2.1379
190000/ 200000: 1.8419
plt.plot(lossi)
[<matplotlib.lines.Line2D at 0x7f0278310110>]
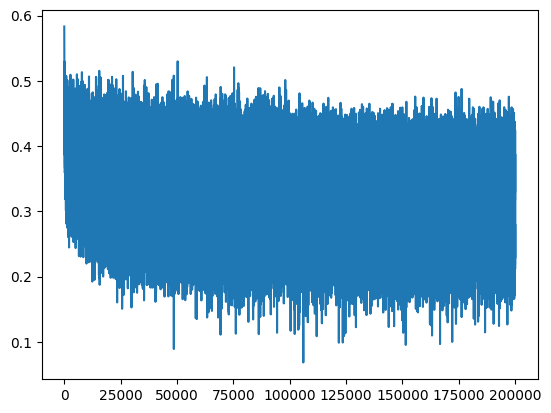
We now have a loss curve that does not start at an aberrant value, which speeds up optimization.
Another Problem#
One might think that a high loss is not necessarily a problem. However, poor weight initialization can cause other issues.
Consider the first iteration of training with values initialized without the 0.01 factor.
C = torch.randn((46, embed_dim))
W1 = torch.randn((block_size*embed_dim, hidden_dim))
b1 = torch.randn(hidden_dim)
W2 = torch.randn((hidden_dim, 46))
b2 = torch.randn(46)
parameters = [C, W1, b1, W2, b2]
for p in parameters:
p.requires_grad = True
ix = torch.randint(0, Xtr.shape[0], (batch_size,))
Xb, Yb = Xtr[ix], Ytr[ix]
emb = C[Xb]
embcat = emb.view(emb.shape[0], -1)
hpreact = embcat @ W1 + b1
h = torch.tanh(hpreact)
logits = h @ W2 + b2
loss = F.cross_entropy(logits, Yb)
for p in parameters:
p.grad = None
loss.backward()
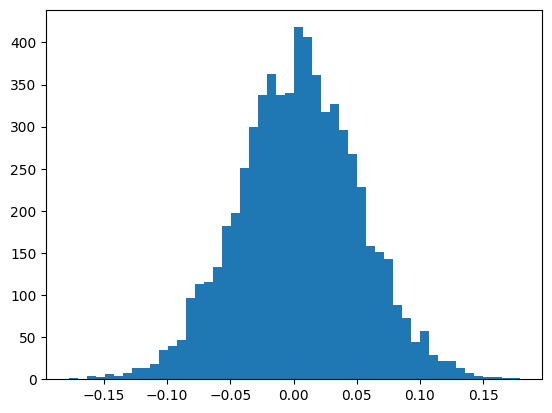
We look at the histogram of values after the tanh activation function.
plt.hist(h.view(-1).tolist(),50);
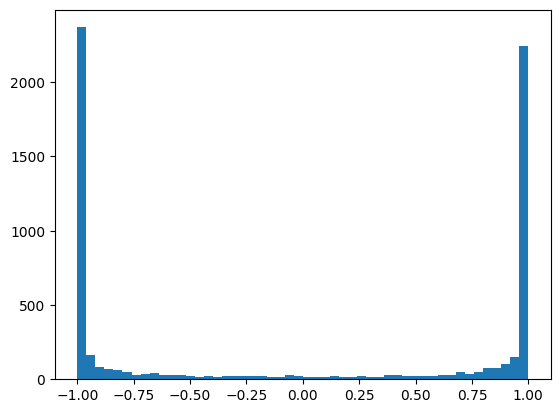
We observe that most of the values are around 1 or -1.
What is the problem with this? When calculating the gradient, using the chain rule, we multiply the gradients of the different calculation steps. The derivative of the tanh function is: \(tanh'(t)= 1 - t^2\) If the values of \(t\) are at 1 or -1, then the gradient will be extremely small (never zero, as it’s an asymptote). This means that the gradient does not propagate, and therefore optimization cannot function optimally at the beginning of training.
We can visualize the values of each neuron.
plt.figure(figsize=(20,10))
plt.imshow(h.abs()>0.99,cmap='gray',interpolation='nearest')
<matplotlib.image.AxesImage at 0x7f02780ae550>
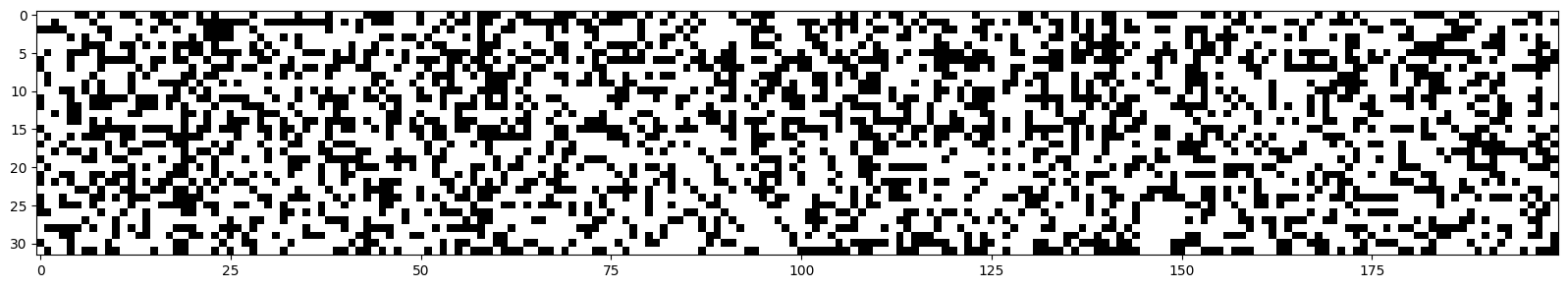
Each white dot corresponds to a neuron whose gradient is approximately equal to 0.
Dead Neuron: If one of these columns were entirely white, it would mean that the neuron does not activate on any element (of the batch). This means it is a useless neuron, which will have no impact on the result and cannot be optimized (on the values present in this batch).
Notes:
This type of behavior is not exclusive to tanh: sigmoid and ReLU can have the same problem.
The problem did not prevent us from training our network correctly, as it is a small model. On deeper networks, this is a big problem, and it is advisable to check the activations of your network at different stages.
Dead neurons can appear at initialization, but also during training if the learning rate is too high, for example.
How to Solve This Problem?#
Fortunately, this problem can be solved in exactly the same way as the problem of the too high loss. To ensure this, let’s look at the activation values and inactive neurons at initialization with our new values.
C = torch.randn((46, embed_dim))
W1 = torch.randn((block_size*embed_dim, hidden_dim)) *0.01# On initialise les poids à une petite valeur
b1 = torch.randn(hidden_dim) *0 # On initialise les biais à 0
W2 = torch.randn((hidden_dim, 46)) *0.01
b2 = torch.randn(46)*0
parameters = [C, W1, b1, W2, b2]
for p in parameters:
p.requires_grad = True
ix = torch.randint(0, Xtr.shape[0], (batch_size,))
Xb, Yb = Xtr[ix], Ytr[ix]
emb = C[Xb]
embcat = emb.view(emb.shape[0], -1)
hpreact = embcat @ W1 + b1
h = torch.tanh(hpreact)
logits = h @ W2 + b2
loss = F.cross_entropy(logits, Yb)
for p in parameters:
p.grad = None
loss.backward()
plt.hist(h.view(-1).tolist(),50);
plt.figure(figsize=(20,10))
plt.imshow(h.abs()>0.99,cmap='gray',interpolation='nearest')
<matplotlib.image.AxesImage at 0x7f025c538190>
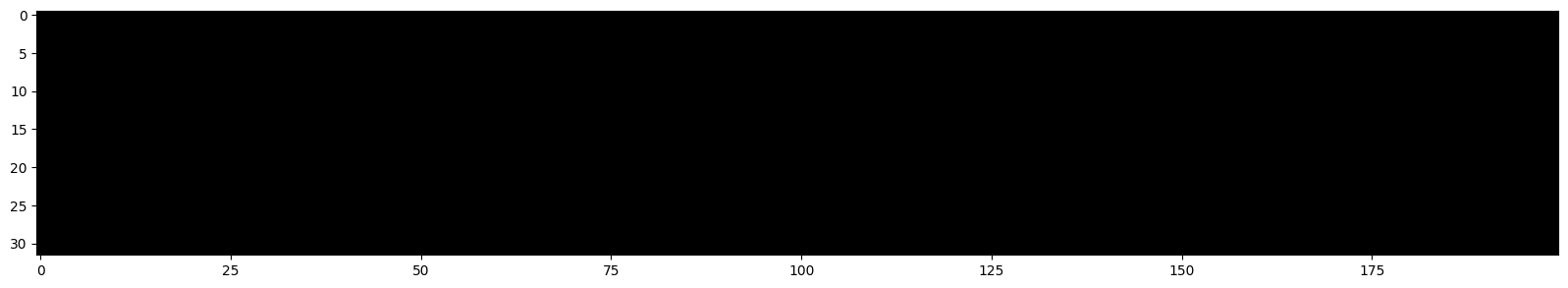
Everything is going well!
Optimal Values at Initialization#
This problem being very important, many researches have been directed towards this subject. A notable publication is Delving Deep into Rectifiers, which introduces the Kaiming initialization. The paper proposes initialization values for each activation function that guarantee a standard normal distribution across the entire network.
This method is implemented in PyTorch, and the layers we will create in PyTorch are directly initialized in this manner.
Why is this course in the bonus section when it seems very important?#
This is indeed a major problem. However, when using PyTorch, everything is already initialized correctly, and it is generally not necessary to modify these values.
Furthermore, many methods have been proposed to mitigate this problem, mainly:
Batch normalization, which we will see in the next notebook, which consists of normalizing the values before activation throughout the network.
Residual connections, which allow the gradient to be transmitted throughout the network without being too impacted by the activation functions.
Despite the importance of these considerations, in practice, it is not always necessary to be aware of them to train a neural network.